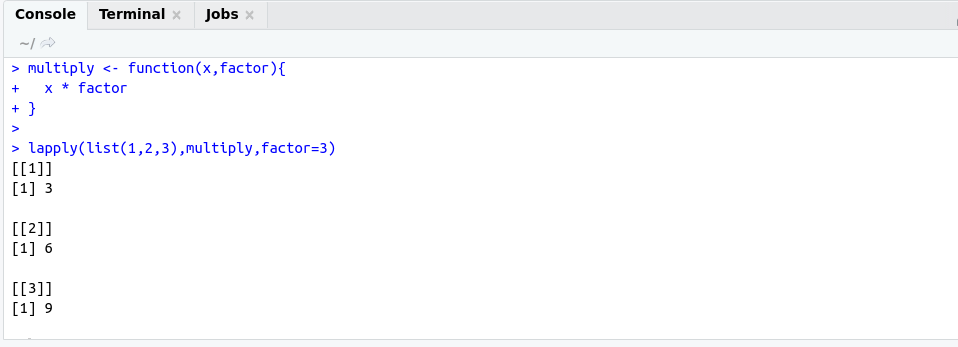
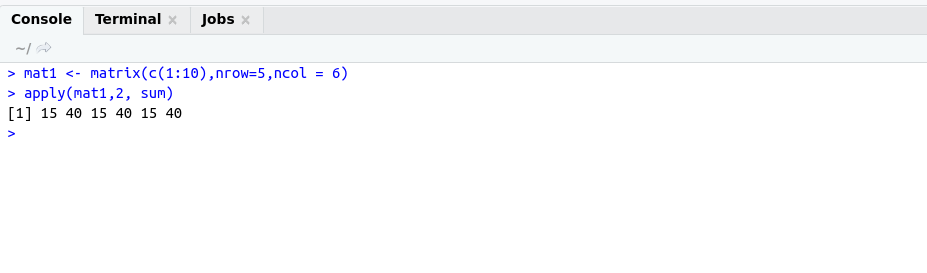
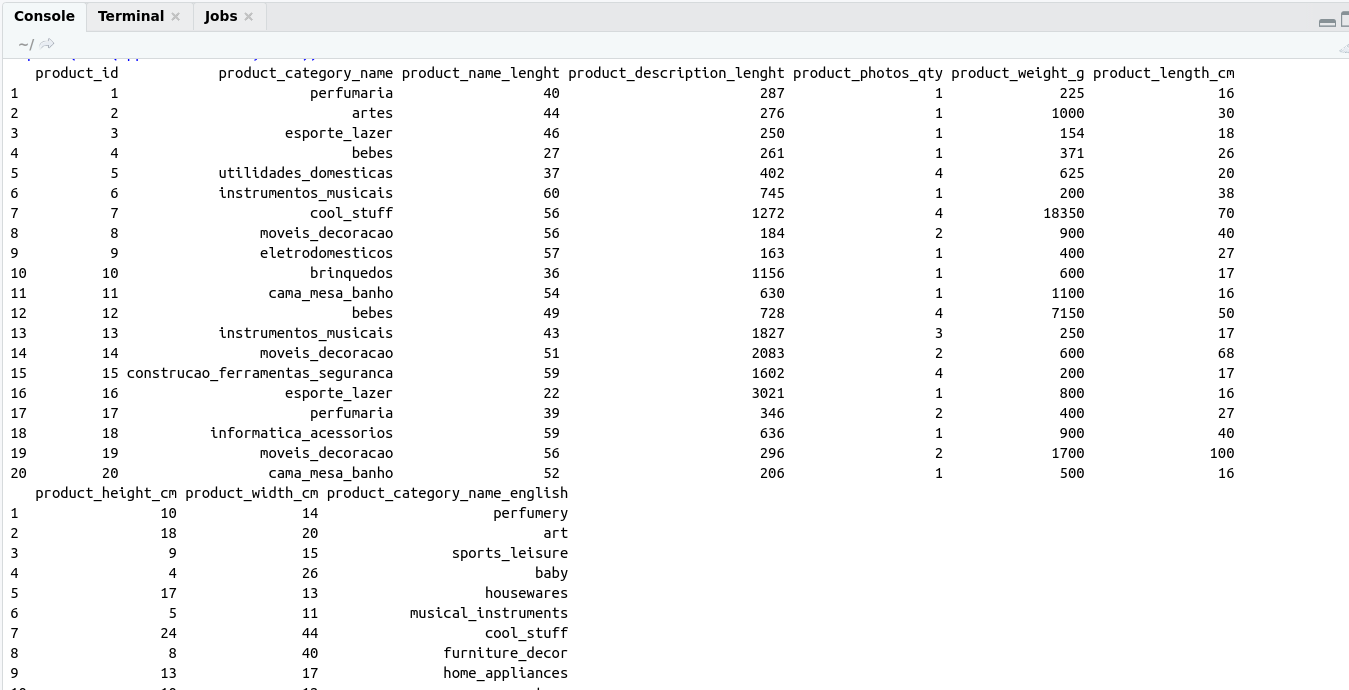
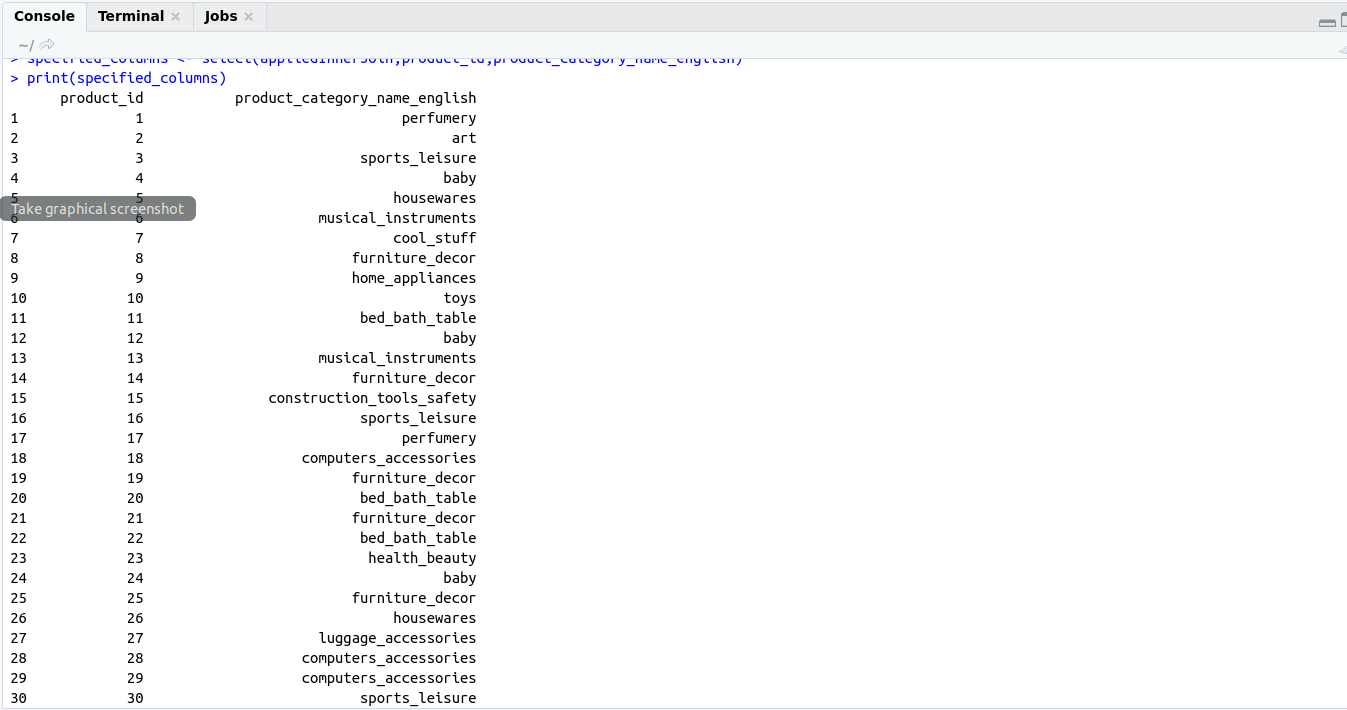
Reactive Programming helps us to build an interactive application using shiny.
In shiny, there are three fundamental components of Reactive Programming :
- Reactive source
- Reactive endpoint
- Reactive conductor
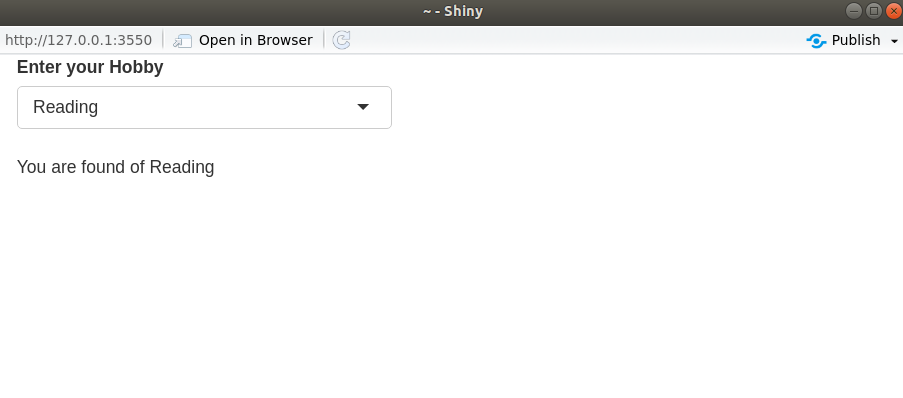
Reactive source
– User input that comes through browser interface typically.
– It can be connected through multiple endpoints.
# load library
library(shiny)
#create ui
ui <- fluidPage(
textInput('name','Enter your name'),
)
# create server
server <- function(input,output,session){
}
# Run the app
shinyApp(ui,server)
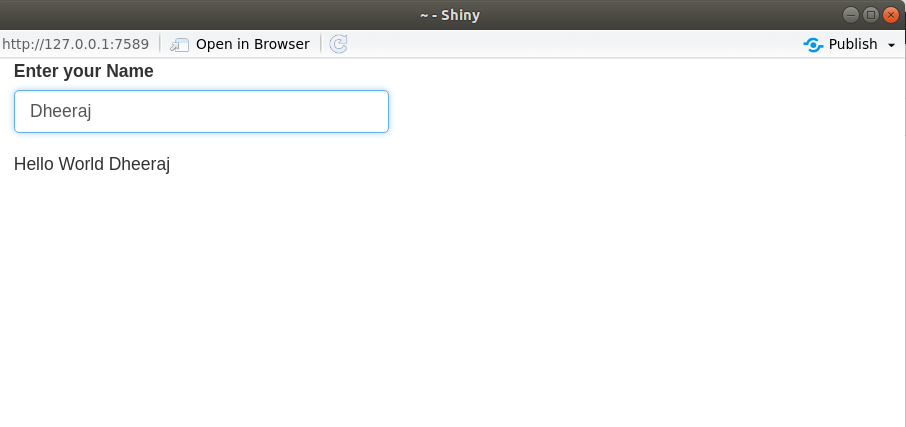
Reactive endpoint
– Something that appears in the user’s browser window, such as a plot or a table of values.
– In simple words, we can say that output that typically appears in the browser window, such as a plot or a table of values.
#load library
library(shiny)
#create ui
ui <- fluidPage(
textInput('name','Enter your name'),
textOutput('greeting')
)
#create server
server <- function(input,output,session){
output$greeting <- renderText({
paste('Hello ',input$name)
})
}
#Run the app
shinyApp(ui,server)
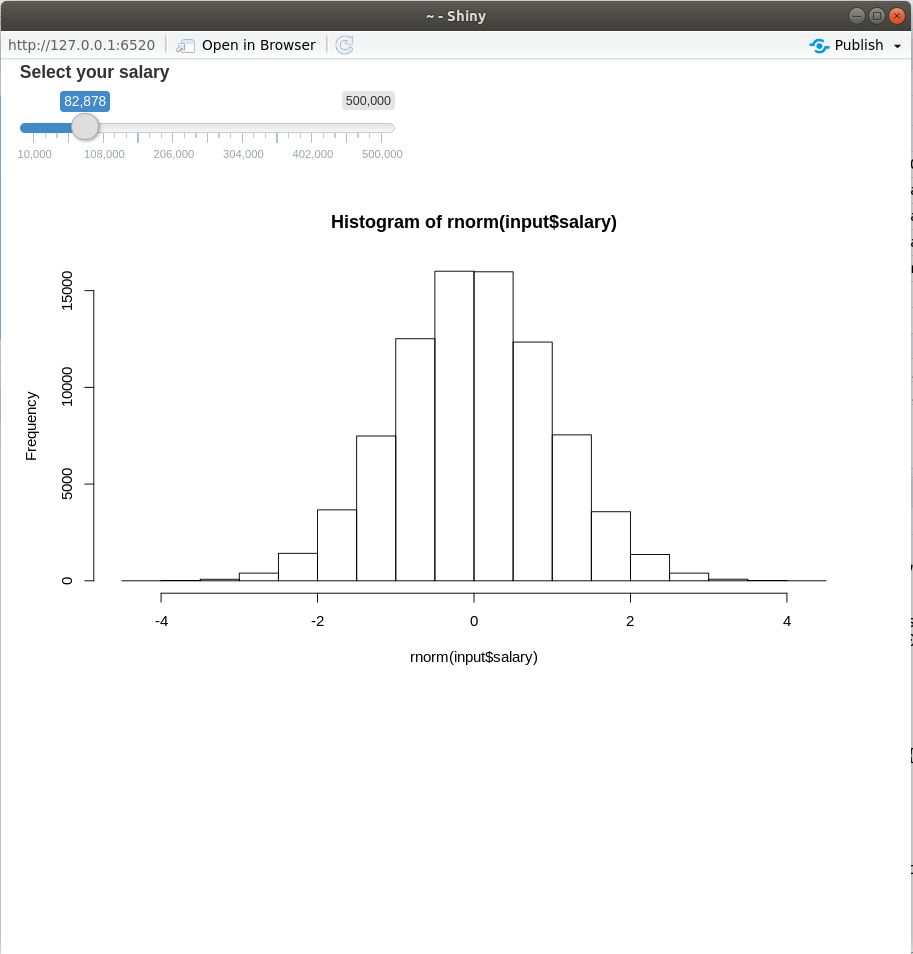
Reactive conductor
– Reactive component between a source and endpoint typically used to encapsulate slow computations.
# create server
server <- function(input,output,session){
#plot putout
output$plot_trendy_names <- ploty::renderPlotly({ babynames %>%
filter(name == input$name) %>%
ggplot(val_bnames, aes(x=year, y=n)) +
geom_col()
})
#table output
output$table_trendy_names <- DT::renderDT({ babynames %>%
filter(name== input$name)
})
}
Reactive Expression
– A Reactive expression is an R expression that uses widget input and returns a value.
– The reactive expression will update this value whenever the original widget changes.
– Reactive expressions are lazy and cached.
To create a reactive expression we use reactive function, which takes an R expression surrounded by braces (just like render function).
ui <- fluidPage(
numericInput('nrows', 'Number of Rows', 10, 5, 30),
tableOutput('table'),
plotOutput('plot')
)
# create server
server <- function(input, output, session){
#input 1
cars_1 <- reactive({
print("Computing cars_1 ...")
head(cars, input$nrows)
})
#input 2
cars_2 <- reactive({
print("Computing cars_2 ...")
head(cars, input$nrows*2)
})
#output plot
output$plot <- renderPlot({
plot(cars_1())
})
#output table
output$table <- renderTable({
cars_1()
})
}
# Run the app
shinyApp(ui = ui, server = server)
Note: A reactive expression can call other reactive expressions.That allows us to modularize computations and ensure that they are NOT executed repeatedly.
You should also visit
Build a Hello world shiny App With R