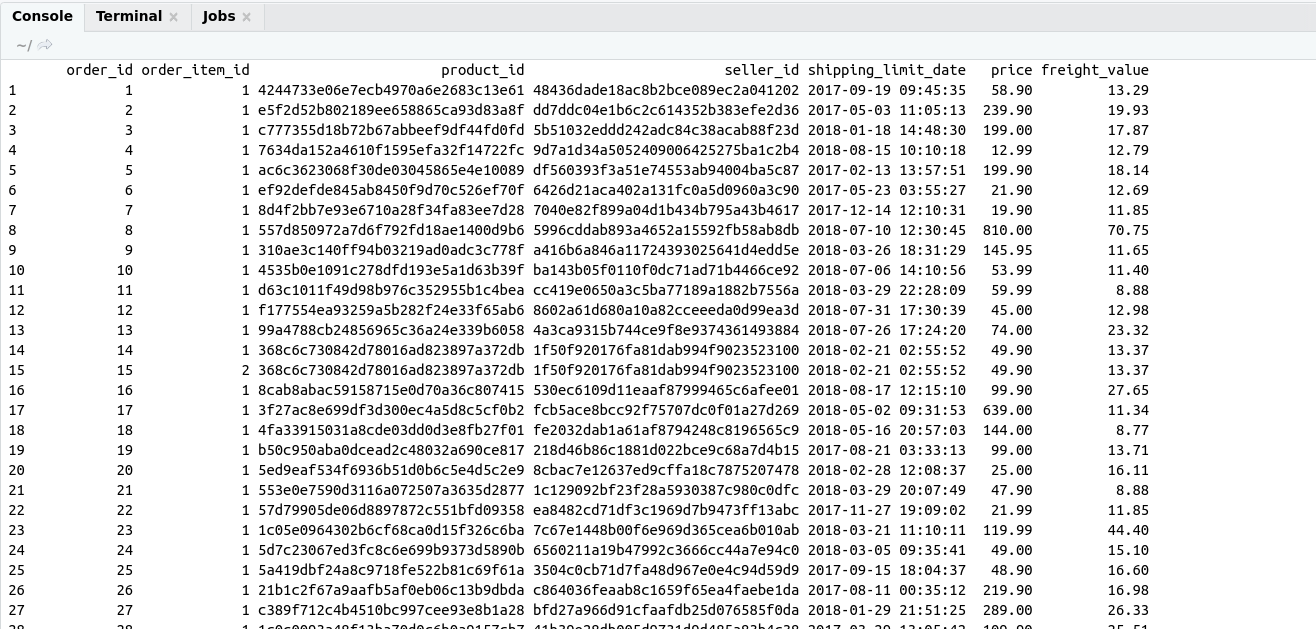
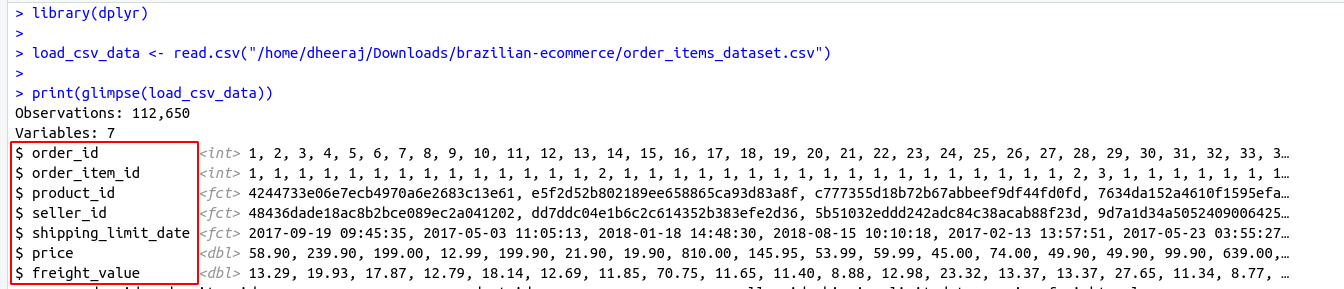
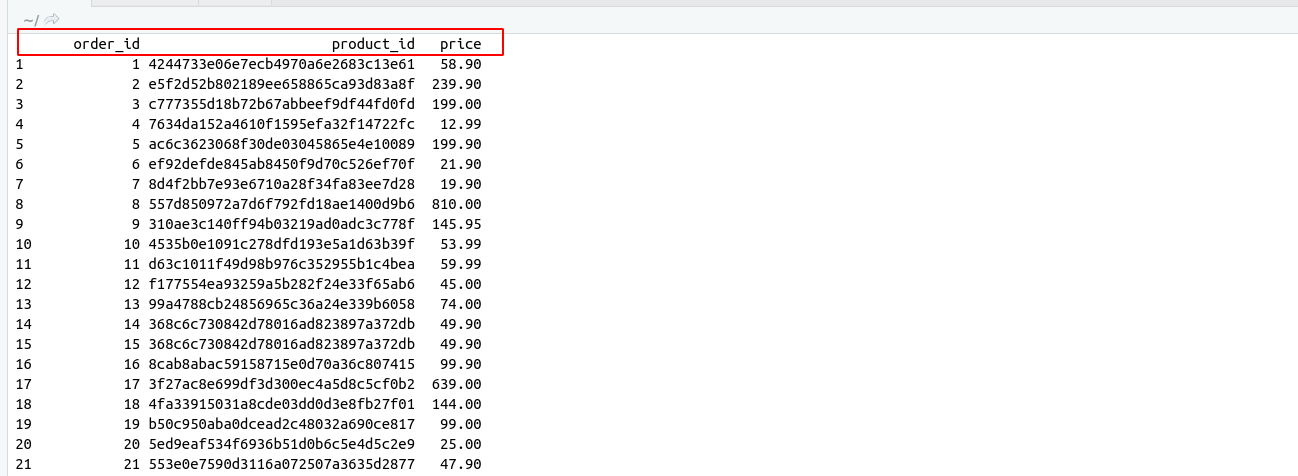
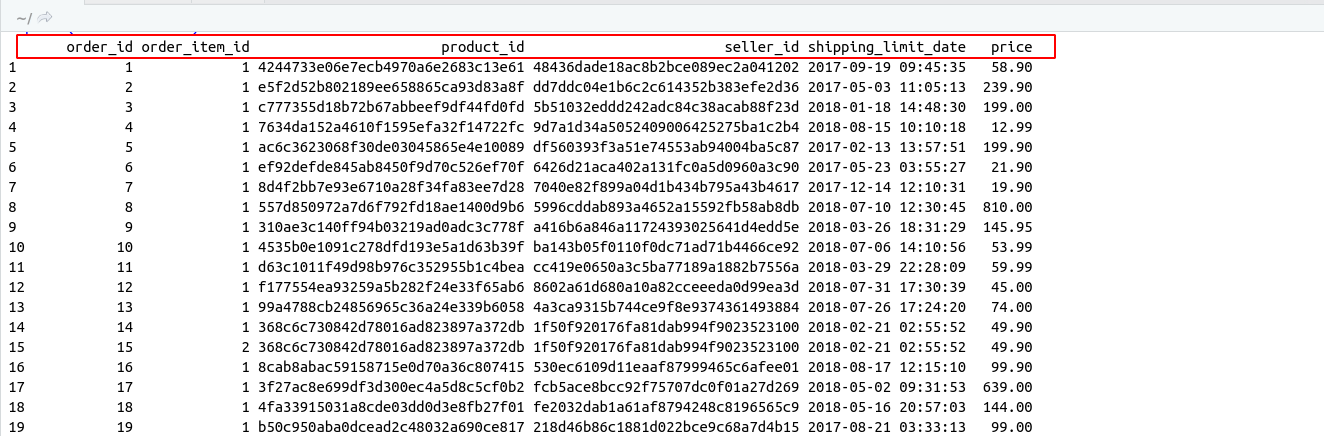
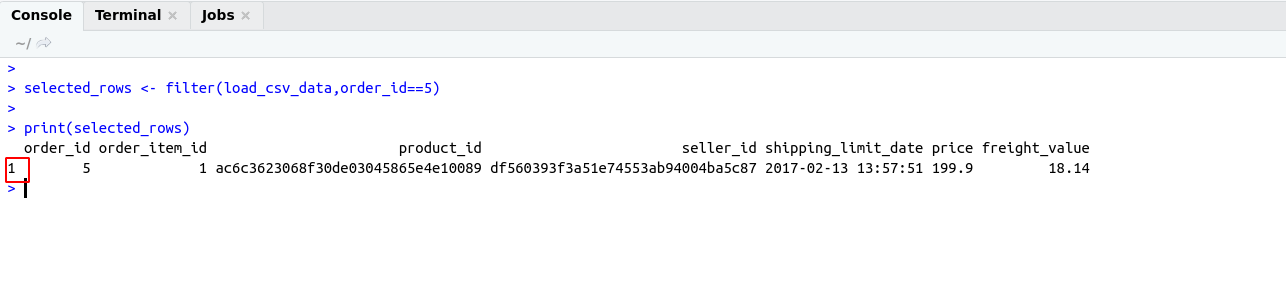
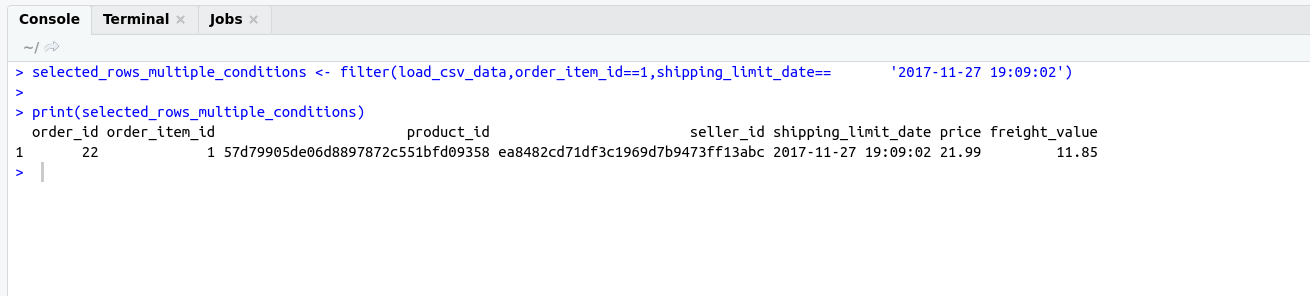
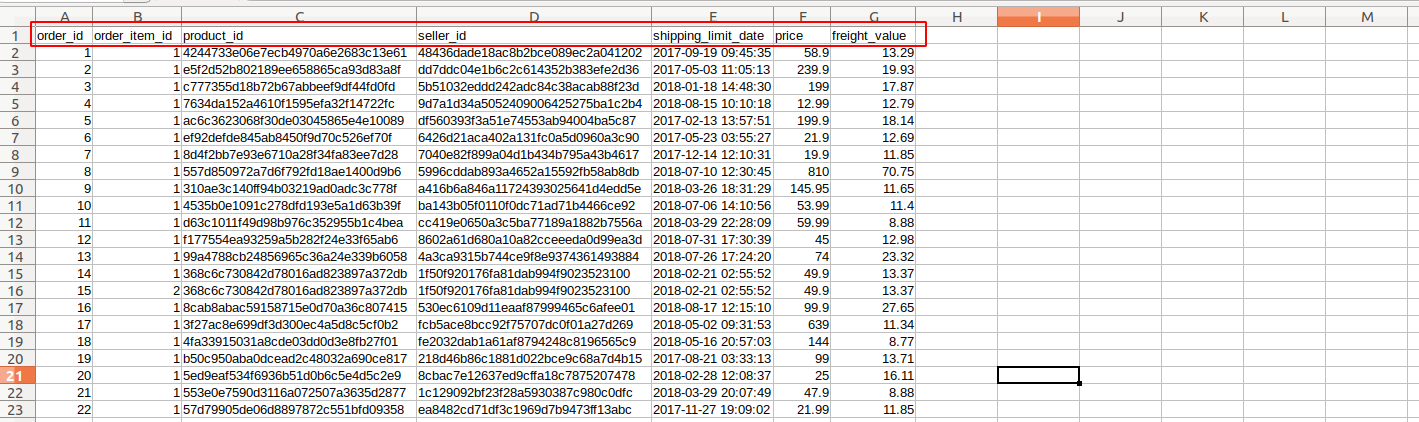
There are some important functions that we are using in Day to day life. I have categories these functions in these categories for understanding.
- Common function
- String Function
- Looping function
Common function
At first, I am dealing with some common functions in R that we use in day to day life during development.
abs() : Calculate absolute value.
Example:
# abs function
amount <- 56.50
absolute_amount <- abs(amount)
print(absolute_amount)
sum(): Calculate the sum of all the values in the data structure.
Example:
# sum function
myList <- c(23,45,56,67)
sumMyList <- sum(myList)
print(sumMyList)
mean() : Calculate arithmetic mean.
Example:
# mean function
myList <- c(23,45,56,67)
meanMyList <- mean(myList)
print(meanMyList)
round() : Round the values to 0 decimal places by default.
Example:
############## round function #####################
amount <- 50.97
print(round(amount));
seq(): Generate sequences by specifying the from, to, and by arguments.
Example:
String Function
# seq() function
# seq.int(from, to, by)
sequence_data <- seq(1,10, by=3)
print(sequence_data)
rep(): Replicate elements of vectors and lists.
Example:
#rep exampleString Function
#rep(x, times)
sequence_data <- seq(1,10, by=3)
repeated_data <- rep(sequence_data,3)
print(repeated_data)
sort(): sort a vector in ascending order, work on numerics.
Example:
#sort function
data_set <- c(5,3,11,7,8,1)
sorted_data <- sort(data_set)Functionround
print(sorted_data)
rev(): Reverse the elements in a data structure for which reversal is defined.
Example:
# reverse function
String Function
data_set <- c(5,3,11,7,8,1)
sorted_data <- sort(data_set)
reverse_data <- rev(sorted_data)
print(reverse_data)
str(): Display the structure of any R Object.
Example:
# str function
myWeeklyActivity <- data.frame(
activity=c("mediatation","exercie","blogging","office"),
hours=c(7,7,30,48)
)
print(str(myWeeklyActivity))
append() : Merge vectors or lists.
Example:
#append function
activity=c("mediatation","exercie","blogging","office")
hours=c(7,7,30,48)
append_data <- append(activity,hours)
print(append_data)
is.*(): check for the class of an R Object.
Example:
#is.*() function
list_data <- list(log=TRUE,
chStr="hello"
int_vec=sort(rep(seq(2,10,by=3),times=2)))
print(is.list(list_data))
as.*(): Convert an R Object from one class to another.
Example:
#as.*() function
list_data <- as.list(c(2,4,5))
print(is.list(list_data))
String Function
Now we are discussing some string function that plays a vital role during data cleaning or data manipulation.
These are functions of stringr package.So, before using these functions at first you have to install stringr package.
# import string library
library(stringr)
str_trim () : removing white spaces from string.
Example:
############### str_trim ####################
trim_result <- str_trim(" this is my string test. ");
print("Trim string")
print(trim_result)
str_detect(): search for a string in a vector.That returns boolean value.
Example:
############### str_detect ####################
friends <- c("Alice","John","Doe")
string_detect <- str_detect(friends,"John")
print("String detect ...")
print(string_detect)
str_replace() : replace a string in a vector.
Example:
############## str_replace #####################
str_replace(friends,"Doe","David")
print("friends list after replacement ....");
print(friends);
tolower() : make all lowercase.
Example:
############## tolower #####################
myupperCasseString <- "THIS IS MY UPPERCASE";
print("lower case string ...");
print(tolower(myupperCasseString));
toupper() : make all uppercase.
Example:
############## toupper #####################
myupperCasseString <- "My name is Dheeraj";
print("Upper case string ...");
print(toupper(myupperCasseString));
Lopping
lapply(): Loop over a list and evaluate a function on each element.
Some important points regarding lapply :
# lapply takes three arguments:
- list X
- function (or name the function) FUN
- … other argumnts
# lapply always returns list, regardless of the class of input.
Example:
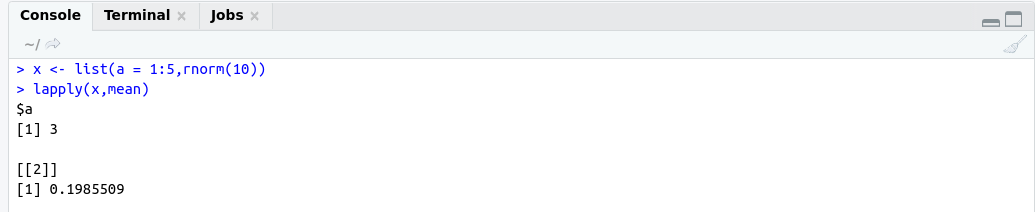
############## lapply example #####################
x <- list(a = 1:5,rnorm(10))
lapply(x,mean)
OutPut:
Anonymous function
Anonymous functions are those functions that have no name.
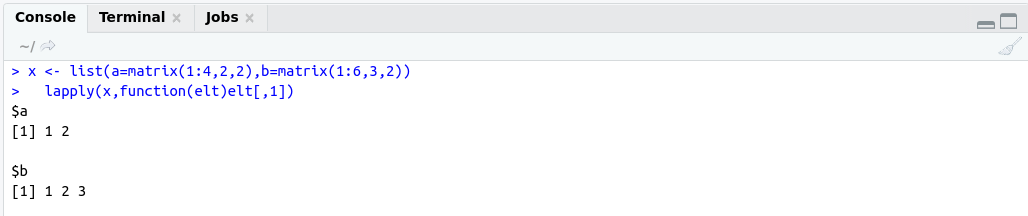
############## lapply example #####################
# Extract first column of matrix
x <- list(a=matrix(1:4,2,2),b=matrix(1:6,3,2))
lapply(x,function(elt)elt[,1])
OutPut:
Use function with lapply
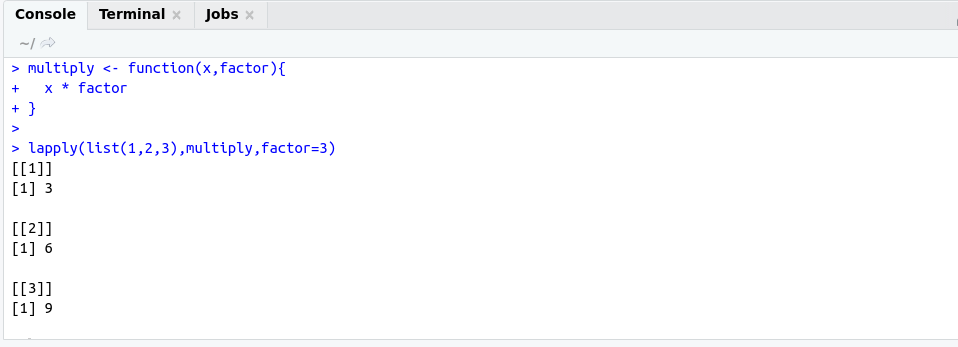
############## lapply example #####################
# multiply each element of list with factor
multiply <- function(x,factor){
x * factor
}
lapply(list(1,2,3),multiply,factor=3)
OutPut:
sapply(): Same as lapply but try to simplify the result.
Example:
############## sapply example #####################
# multiply each element of list with factor
multiply <- function(x,factor){
x * factor
}
sapply(list(1,2,3),multiply,factor=3)
OutPut:

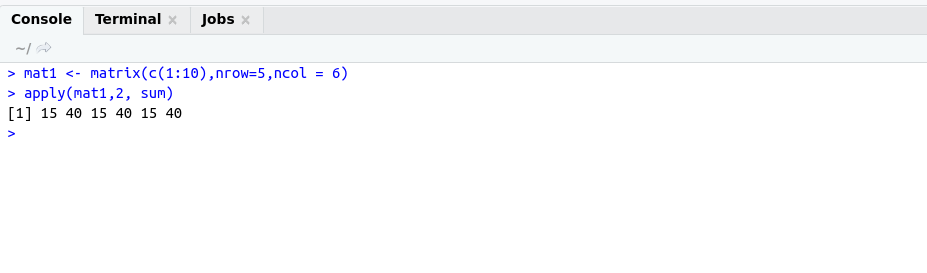
apply() : Apply a function over the margin of an array.
Example:
############## apply function #####################
mat1 <- matrix(c(1:10),nrow=5,ncol = 6)
apply(mat1,2, sum)
OutPut:
tapply(): Apply a function over subsets of a vector.
Example:
############## tapply function #####################
tapply(mtcars$mpg, list(mtcars$cyl, mtcars$am), mean)
OutPut:
mapply():Multivariate version of lapply.
Example: